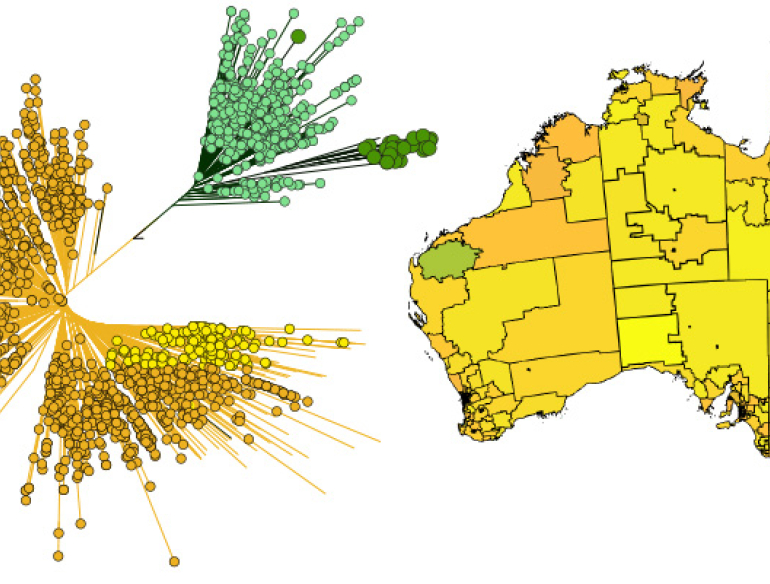
Australia is at the forefront of successful control of human immunodeficiency virus type 1 (HIV-1) transmission due to rapid and comprehensive public health responses from the beginning of the epidemic in the 1980s. In that regard, Australia has recently experienced its first major decline in new HIV-1 infections in ten years, and which is attributed to the most recent public health intervention approach named pre-exposure prophylaxis (PrEP). However, there are state-specific differences; in 2018 New South Wales (NSW) reported a 21% but South Australia (SA) only a 0.8% decline in new HIV-1 notifications compared to the 5-year average. Also, NSW reported a decline in new notifications of 41% for Australian-born men who have sex with men (MSM), but only 6% for non-Australian-born MSM between 2014–2018.This is indicative of changes in the epidemic as infected populations are becoming increasingly heterogeneous. However, without sequence data it is difficult to capture and interpret the changes in transmission within and between risk groups.
Our laboratory has established a state-wide database harboring annotated sequence data from 2004 onwards. The data has been successfully used to investigate the changes in transmission over time for specific HIV-1 genetic variants (subtypes) and differences in different transmission risk groups. The main objective is to combine viral evolution with epidemiology to develop methods for tracking transmission in real-time and how to translate these findings to adaptable and effective public health interventions.
NSW harbors a state-wide database that links HIV-1 molecular data with demographic data. This HIV-1 linkage-database will be the source of data for the proposed project. It currently contains approximately 8,000 HIV-1 sequences from the past 14 years and will include prospective data collected annually. Over 70% of sequence data contains linked demographic data, and which makes this database highly representative of the true virus population in NSW. Tools used will include alignment programs (Geneious), phylogenetic tools (IQ Tree, FastTree), and computational analysis tools (Matlab, R).
We have identified major differences in transmission dynamics between subtype B (most common subtype in Australia) and non-B subtypes infections. Demographics regarding region born, transmission risk factor, stage of infection, and geographic location differ between subtypes. In addition, large active transmission clusters have been identified for subtype B and these might be drivers of the local epidemic.
Phylogeny is the study of the relationship of different organisms in past and present, i.e. the evolutionary history. Phylodynamic refers to the study of combined phylogenetic and epidemiological data and its implementation has revolutionized the study of virus emergence. Using a combination of sequence data and epidemiological data will improve the surveillance systems' and improve the response on the epidemic with the ultimate aim of being the first country to virtually eliminate HIV-1 transmission.
- Dr Angie Pinto, Royal Prince Alfred Hospital, Sydney
- NSW HIV Prevention Partnership Project